|
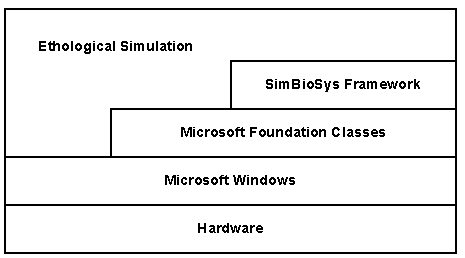
A schematic showing the layers in a simulation application are shown in Figure 20.

|
#define ctor /**/ #define dtor virtual #define override virtual
Figure 21. Pseudo-keywords
The first two, ctor and dtor, indicate a constructor and
destructor respectively. The pseudo-keywords take the place of return types in
the header allowing all the members names to line up nicely in a column. The
ctor keyword is a placeholder, being defined as just an open and close comment.
Defining dtor as virtual ensures that deleting an instance of the class will
call the correct destructor even if the pointer type refers to one of the
class's base classes. The override symbol indicates that the member
function is overloading one of its base class's member functions.
Framework
Classes
This section will present and discuss the implementations of the classes
designed in the previous chapter. All of the `top-level' SIMBIOSYS framework
classes have been derived from the MFC base class CObject. This policy has two
advantages. The MFC framework provides several collection classes which work
with instances of classes derived from CObject. The linked list CObList is
particularly useful. Secondly, CObject has facilities for persistence built in
which all derived classes can exploit.

|
class bioSimulation : public bioObject
{
public:
ctor bioSimulation (void);
dtor ~bioSimulation (void);
virtual void SetWorld (bioWorld*);
virtual bioWorld* GetWorld (void);
virtual void AddPopulation (bioPopulation*);
virtual int GetData (int src, void* dest);
virtual int NextStep (void);
virtual int StartEnvCycle (void) {return FALSE;}
virtual int StartBreedCycle (void) {return FALSE;}
virtual void DoBreedCycle (void);
virtual void DoEnvCycle (void);
virtual void DoActionCycle (void);
protected:
bioWorld* _world;
CObList _populations;
int _curAct;
int _totalAct;
int _curEnv;
int _totalEnv;
int _curGen;
};
Figure 23. bioSimulation class header
The simulation relies on an external source to trigger clock events through the NextStep() method. This would typically be done from a "Single Step" menu item handler and the handler of a system timer which is started and stopped through menu events. The frequency of the timer is dependent on several factors: the amount of computation that must be done in an action cycle, the amount of redrawing for a single frame in the map animation, and the hardware platform. Typically, 20 frames per second (a 50 ms time interval for the timer) is pushing the limits of the simulation.
The NextStep() method first calls StartEnvCycle() to see if an environment cycle should be executed. If this returns true, StartBreedCycle() is called to see if a breeding cycle should be executed. If the latter also returns true, DoBreedCycle() is called before DoEnvCycle(). In any case, DoActionCycle() is called at the end. StartEnvCycle() and StartBreedCycle() both default to returning false, so the default behavior of NextStep() is to only call DoActionCycle() which sends NextStep() to the world instance. The default implementation of DoBreedCycle() sends the Breed() message to all populations stored in the simulation and increments the internal counters related to breeding cycles. The default implementation of DoEnvCycle() only increments the internal counters. If an application wants to make use of breeding cycles and/or environment cycles, it will have to override the Start*Cycle() methods and return true when certain conditions are met such as after a certain number of action cycles.
The GetData() method provides a single interface for all instrument objects and
other interested clients to retrieve information from the simulation. The type
of data is specified by a value passed in with the src variable. If the
simulation recognizes the src, it will set the dest pointer to whatever the src
parameter specifies, otherwise it will pass the request to its base class. In
this way, subclasses of bioSimulation can easily extend the types of data
provided without expanding the public interface.
bioWorld
The environment is implemented in the bioWorld abstract base class. Its purpose
is to keep track of all the things in the environment, their position and
orientation, and to oversee the redrawing of the map in the user interface. It
also manages the action cycles, giving each thing in the world a turn to
perceive its local environment and determine an intention. The world is
responsible for resolving each thing's intention into an action, though
subclasses of bioWorld may delegate that responsibility to other classes.
class bioWorld : public bioObject
{
public:
ctor bioWorld (void);
dtor ~bioWorld (void);
virtual void Init (void) {}
virtual void AddThing (bioThing*, double x, double y);
virtual void RemoveThing (bioThing*);
virtual void NextStep (void);
virtual void ResolveAction (bioThing*, int intent) = 0;
virtual void PlaceRandom (bioThing*) = 0;
virtual void MoveThing (bioThing*, double x, double y,
int relative = 0);
virtual void TurnThing (bioThing*, double angle,
int relative = 0);
virtual bioObject* WhatsAt (double x, double y, bioThing*) = 0;
virtual void XformPos (double x, double y, bioThing*,
double& nx, double& ny);
virtual void Redraw (CDC*) = 0;
virtual void Resize (int width, int height) = 0;
virtual void UserSelect (int x, int y) {}
virtual void UserEdit (int x, int y) {}
protected:
void _SetPosition (bioThing*, double x, double y);
CObList _things;
};
Figure 24. bioWorld class header
Init() is a virtual method which allows subclasses to do any initialization best left out of their respective constructors such as memory allocation. AddThing() and RemoveThing() are public interfaces to the bioWorld's internal list of bioThing instances. An initial position must be provided with AddThing(). If an initial position is not known, the client should call PlaceRandom() which will find an empty spot on the world for the new thing. It is the responsibility of subclasses of bioWorld to implement PlaceRandom() because there is not enough information stored in bioWorld or bioThing to ensure that new things won't overlap or even if overlapping is allowed.
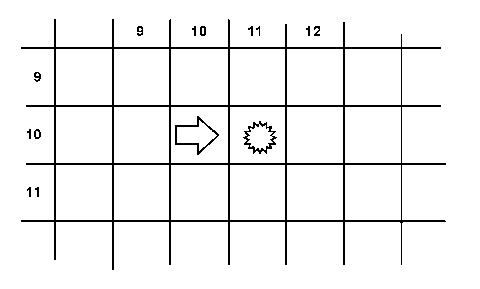
NextStep() executes a single action cycle. Each bioThing instance in the world's list is sent the Look() message which gives the thing a chance to perceive its local environment by sending the world one or more WhatsAt() messages. The WhatsAt() method returns the contents of the position provided by the thing, relative to the thing's current position and orientation. For instance, if a thing is at location (10, 10) and has a heading of 90 and asks what's at (0, 1), i.e. what is directly in front me, then the world would return the contents of location (11, 10) as shown in Figure 25. This is accomplished partially through the use of XformPos(), a method which transforms a position specified by an x- and y-coordinate into a different coordinate system. Separating XformPos() out into its own virtual method allows subclasses to optimize the transformation. The default implementation is quite general, and its use of transformation matrices is relatively computationally intensive.
After all things have been told to Look(), each thing is queried for its intention, returned as an integer value, which is passed to ResolveAction(). This is a pure virtual function, therefore subclasses must provide an implementation which determines how intentions are resolved into actions, that is, how the interaction of what all the things want to do is manifested into actual events.
MoveThing() and TurnThing() are used to manipulate the location and orientation of a thing respectively. Both methods take an extra parameter which specifies whether the change is absolute or relative to the thing's current state. The parameter is optional and defaults to absolute. The _SetPosition() protected method allows subclasses of bioWorld to set the position of a thing directly without having to first add it to the list of things.
The final four methods have to do with the user interface. Redraw() is responsible for drawing a single frame of the map animation into the passed in device context. Resize() is used to inform the world instance that the area it draws into has changed. UserSelect() and UserEdit() are to be called when the user clicks and double-clicks on the map. Subclasses are responsible for converting the device coordinates of the mouse event into world coordinates, finding the thing instance at that location, and sending that thing Select() and Edit() respectively.
|
class bioCellWorld : public bioWorld
{
public:
ctor bioCellWorld (void);
dtor ~bioCellWorld (void);
virtual void SetSize (int);
virtual void ReadMap (char*);
virtual void CreateThing (char id, int x, int y) {}
override void AddThing (bioThing*, double x, double y);
override void RemoveThing (bioThing*);
override void PlaceRandom (bioThing*);
override bioObject* WhatsAt (double x, double y, bioThing*);
override void Resize (int width, int height);
override void UserSelect (int x, int y);
override void UserEdit (int x, int y);
protected:
int _size; // how many cells across
int _width; // how many pixels across one cell
int _side; // how many pixels across the world
int _winWidth; // window width
int _winHeight; // window height
int _offX; // horizontal offset
int _offY; // vertical offset
bioThing*** _cells; // discrete regions of the world
};
Figure 26. bioCellWorld Class Header
The world is initialized using one of two methods. SetSize() will create
an empty world of the specified size. ReadMap() will read an ASCII file
containing a representation of the world. The first line contains the size.
There should follow a number of lines equal to size, each containing size
characters. For each character in the file, the method CreateThing() is called
along with the position of that character. Subclasses of bioCellWorld are
expected to override this method and create an appropriate bioThing instance
represented by that character. If it doesn't recognize the character, it should
invoke its base class's implementation.
bioThing
bioThing provides the bioWorld class a public interface for all objects
that can inhabit a world. Even though subclasses of bioThing can be completely
passive (e.g. rocks and food), the bioThing class must define virtual functions
for looking and retrieving intentions so that the world has a way to send
messages to the active objects.
class bioThing : public bioObject
{
public:
ctor bioThing (void);
dtor ~bioThing (void);
virtual int ClassID (void) {return 0;}
bioWorld* GetWorld (void) {return _world;}
double GetX (void) {return __x;}
double GetY (void) {return __y;}
double GetOrientation (void) {return __orientation;}
virtual void Look (void) {}
virtual int GetIntent (void) {return 0;}
virtual void Select (void) {}
virtual void Edit (void) {}
friend class bioWorld;
protected:
bioWorld* _world;
private:
double __x;
double __y;
double __orientation;
};
Figure 27. bioThing class header
ClassID() provides run-time class information in the form of an integer value. This is useful to the world instance if it wants to do all the drawing and intention resolution rather than delegating those tasks to the things themselves. GetWorld(), GetX(), GetY(), and GetOrientation() simply return the current values stored in the instance.
Look() is sent by the world once for every action cycle. This gives the thing an opportunity to query the world for the current state of its local environment before it decides on an intention. The default implementation of Look() does nothing and GetIntent() returns 0 which should be interpreted as an intent to do nothing. Unless a subclass overrides both, it will be a passive object.
Likewise, Select() and Edit() are no-ops at this level. It is up to subclasses
to define what should happen if the user clicks and double-clicks on a thing.
bioAgent
A bioAgent is a thing whose behavior is driven by an internal program.
class bioAgent : public bioThing
{
public:
ctor bioAgent (void);
dtor ~bioAgent (void);
void SetProgram (bioProgram*);
bioProgram* GetProgram (void);
override void Look (void) = 0;
override int GetIntent (void) {return _intention;}
protected:
bioProgram* _program;
int _intention;
};
Figure 28. bioAgent class header
SetProgram() and GetProgram() simply store and retrieve the agent's
internal program instance. Look() has been redefined as a pure virtual which
forces subclasses to provide an implementation. Subclasses are expected to set
the value of the protected _intention data member during their Look() which is
returned through bioAgent's implementation of GetIntent().
bioProgram
The bioProgram abstract base class provides a minimal public interface
for the bioAgent class.
class bioProgram : public bioObject
{
public:
virtual void Init (void) {}
virtual int Think (int* inputVector, int size) = 0;
protected:
};
Figure 29. bioProgram class header
The Init() method allows subclasses to do any initialization best left
out of the constructor such as allocating memory. The agent which owns the
program will query its local environment during its Look() method. The
information it retrieves from the environment is translated into an input
vector in the form of an array of integers which is passed to the program's
Think() method. Subclasses of bioProgram should use this input vector as input
to whatever program they implement and return an intention in the form of an
integer value.
bioFSM
bioFSM implements a finite state machine program. The FSM is a look-up
table which combines it current state and the input into an index. The contents
of the table at the index yields an output and the next state of the table.
class bioFSM : public bioProgram
{
public:
ctor bioFSM (int, int, int);
dtor ~bioFSM (void);
override void Init (void);
override int Think (int* inputVector, int size);
virtual void ReadArray (int* array, int size);
protected:
int _state;
int _stateSize;
int _stateMask;
int _inputSize;
int _inputMask;
int _output;
int _outputSize;
int _outputMask;
int* _table;
int _tableSize;
int _index;
};
Figure 30. bioProgram class header
The constructor requires three parameters: the number of bits in the state, input and output. Init() calculates the derived data members and allocates space for the table. ReadArray() populates the table from the passed in array and is provided to allow agents to set the table from an instance of bioGType().
Think() expects an input vector with a size equal to the input size specified
in the constructor. It constructs an index from its current state and the input
vector, looks up a new state, and returns the output defined in the table at
that index.
bioNNet
The bioNNet class implements a recurrent neural network. Each node in
the network is connected to every other node by an integer weight. A zero
weight is equivalent to not being connected. At any given time a node can be in
one of two states, on or off. Nodes are designated as being input, internodes
or output. The only difference between them is that the state of input nodes
are determined outside the network, and the state of output nodes is exported
outside the network.
class bioNNet : public bioProgram
{
public:
ctor bioNNet (int, int, int);
dtor ~bioNNet();
override int Think (int* vector, int size);
virtual void ReadArray (int* input, int count);
void SetSize (int, int, int);
protected:
void _FireNeuron (int, int);
int _IsOutput (int);
int _Compute();
int _size;
int _inputs;
int _inters;
int _outputs;
int* _neuron;
int* _newValues;
int* _base;
int** _weights;
};
Figure 31. bioNNet Class Header
Methods
The number of input nodes, internodes, and output nodes are defined in
the constructor while SetSize() does the actual work of allocating memory and
initializing the values. ReadArray() initializes the values for the weights,
node biases and initial activations. This would be called by bioPType instance
after extracting the information from a bioGType instance. Think() uses the
input vector to set the activations of the input nodes using _FireNeuron(),
calls _Compute which runs the network through a cycle, and returns the states
of the output nodes compressed into a single integer. During a _Compute()
cycle, the inputs to each node are multiplied by their corresponding weight,
summed up and added to the node's bias value and compared to a preset
threshold. If the final sum of the inputs is greater than 100 (chosen
arbitrarily) then the node is turned on (its value is set to 1), otherwise it
is turned off (its value is set to 0).
bioGType
The Genotype object designed in the previous chapter is implemented in
the bioGType abstract base class. Instances are typically owned by instances of
bioPtype and manipulated by instances of bioPopulation. bioPTypes would use an
instance of bioGType to construct the phenotype including their respective
program instances.
class bioGType : public bioObject
{
public:
ctor bioGType (void) {}
dtor ~bioGType (void) {}
virtual void SetLength (int) = 0;
virtual int GetLength (void) = 0;
virtual void Randomize (void) = 0;
virtual void Mutate (double rate) = 0;
virtual bioGType* Crossover (bioGType*) = 0;
protected:
};
Figure 32. bioGType class header
All methods are defined as pure virtuals which forces subclasses to provide implementations. The length of the genotype which defines the number of alleles it stores is set and retrieved via the SetLength() and GetLength() member functions. Randomize() should set all alleles in the genotype to random values which is only useful for the first generation in a population.
For each bit in the genotype, Mutate() should toggle it with the probability
passed in. Crossover() should create a new genotype of the same class by
applying the crossover operator to itself and the instance passed in.
bioHapGType
A haploid (single-strand) genotype is implemented in the bioHapGType
class. It provides implementations for all the pure virtual functions defined
in bioGType.
class bioHapGType : public bioGType
{
public:
ctor bioHapGType (void);
ctor bioHapGType (bioHapGType&);
dtor ~bioHapGType (void);
bioHapGType& operator= (const bioHapGType& rhs);
override void SetLength (int);
override int GetLength (void);
override void Randomize (void);
override void Mutate (double rate);
override bioGType* Crossover (bioGType*);
virtual char GetByte (int idx);
virtual int GetBit (int idx);
virtual void SetBit (int idx, int on = 1);
virtual void ResetBit (int idx);
virtual void ToggleBit (int idx);
protected:
int _bits; // bit count
int _bytes; // string length
char* _dna; // bit string
};
Figure 33. bioHapGType class header
SetLength() allocates space for the genotype and sets the size data
members. GetLength() returns the number of bits. Mutate() doesn't roll the dice
for each bit, but rather uses the mutation rate to calculate the index of the
next bit to be toggled. This method provides substantial performance gains. The
Crossover() method treats the bit strings as circular, choosing two points
randomly on one string and replacing the segment in between with the
corresponding segment from the second bit string. The rest of the methods are
self-explanatory.
bioPType
The Phenotype object designed in the previous chapter is implemented in
the bioPType abstract base class. Phenotypes are agents which are created from
genotypes. Instances of bioPopulation are collections of bioPType instances.
class bioPType : public bioAgent
{
public:
ctor bioPType (bioGType* gtype);
dtor ~bioPType (void);
virtual bioGType* GetGType (void);
virtual bioGType* ExtractGType (void);
virtual double GetFitness (void);
protected:
bioGType* _gtype;
double _fitness;
};
Figure 34. bioPType class header
GetGType() and GetFitness() simply return the instance's current values.
bioPTypes will delete their _gtype member in the destructor. ExtractGType() is
used by bioPopulations to transfer a bioGType instance from one phenotype to
another, essentially a cloning operation.
bioPopulation
The Population object is realized in the bioPopulation abstract base
class. Instances of bioPopulation store and manipulate and collection of
bioPType instances along with their corresponding bioGType instances.
bioPopulations essentially implement the genetic algorithm.
class bioPopulation : public bioObject
{
public:
ctor bioPopulation (void);
dtor ~bioPopulation (void);
virtual void Breed (void);
virtual void SetSize (int);
virtual int GetSize (void);
virtual void SetMutationRate (double);
virtual double GetMutationRate (void);
virtual bioGType* GenGType (void) = 0;
virtual bioPType* GenPType (bioGType*) = 0;
virtual bioPType* GetPType (int idx);
virtual double GetMaxFitness (void);
virtual double GetMinFitness (void);
virtual double GetAvgFitness (void);
protected:
bioGType* _ChooseParent (void);
int _size;
int _elite;
bioPType** _ptypes;
double _mutationRate;
double _totalFitness;
double _maxFitness;
double _minFitness;
double _avgFitness;
};
Figure 35. bioPopulation
SetSize() allocates space for the population and generates an initial population by calling GenGType() and GenPType(). GetSize() returns the current population size. The population's mutation rate is stored and retrieved with SetMutationRate() and GetMutationRate() respectively.
Breed() performs a single step in the genetic algorithm. The population array is sorted according to fitness. Minimum, maximum, and total fitness are recorded and average fitness is calculated. Space for the next generation is allocated and the elite phenotypes are created with GenPType() using genotypes directly from the current generation. The rest of the next generation is created by following these steps: